The importance of microbiomes to the health of their host plants and animals is well known, but the sensitivity and resilience of microbiomes to environmental changes are less understood. At Oregon State University, a multidisciplinary team aims to unlock unexplored realms of these complex communities of microorganisms. With support from the National Science Foundation, this convergence of experts from diverse disciplines shows potential to create industry collaborations and groundbreaking advances in fields such as pharmaceutical discovery and environmental conservation, and to influence developments in personalized medicine and ecological sustainability.
Led by Maude M. David, an expert in microbial ecology, computational biology, and artificial intelligence, and Xiaoli Fern, a specialist in AI across multiple disciplines, the team includes three other distinguished scientists — Rebecca Vega Thurber, Ryan Mueller, and Thomas Sharpton — each of whom possess extensive knowledge of the specific lifeforms under study. Their focus is the unknown processes through which microbiomes adapt to environmental changes and the resulting impacts on the health of their hosts.
The microbiome project in a nutshell
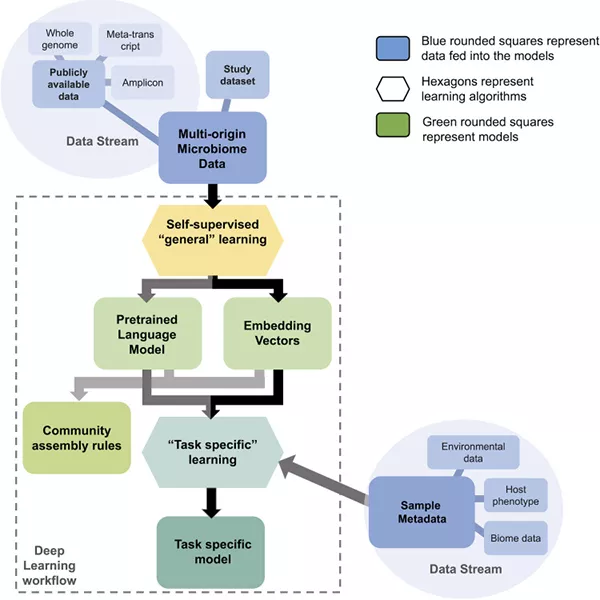
The project started in 2020 and is set to conclude in 2025. Operationally, the team is focused on disturbances impacting the microbiomes of three aquatic organisms: seagrasses, corals, and zebrafish.
By studying the interaction of microbiomes with three environmentally relevant stressors — antibiotics, warming waters, and pathogenic infection — the project aims to resolve the universal properties of microbiomes that influence stress responses. The objective is to develop modeling approaches and System Agnostic Microbiome Measures involving common ecological metrics and novel ones developed using AI algorithms. These will allow for the viewing of microbial characteristics independent of any specific system.
As this exploration unfolds, a web of scientific inquiry emerges, where each piece, brought by distinguished researchers, adds depth and dimension to the comprehensive understanding of microbial ecosystems.
Rebecca Vega Thurber, Emile F. Pernot Distinguished Professor in Microbiology, created a detailed examination of microbial interactions in reef ecosystems provides insights into the adaptive mechanisms and symbiotic relationships intrinsic to corals, unearthing the foundational elements contributing to coral health and resilience.
Ryan Mueller, associate professor of microbiology, contributed his understanding of marine microbial ecology, focuses on unraveling the metabolic pathways and biogeochemical cycles within seagrass ecosystems. His study elucidates the metabolic interactions and ecological roles of microbes in maintaining seagrass health, a crucial component in oceanic carbon sequestration processes.
Thomas Sharpton, associate professor of microbiology, applied his nuanced research in zebrafish microbiomes to decoding the molecular dialogues between the host and its resident microbes. Through intricate analysis of zebrafish as a model organism, he is dissecting the complex dynamics of host-microbe interactions and the resultant physiological alterations, which provide a model for understanding similar interactions in humans.
Analytical depth and technological integration
As the data is collected, the integration of advanced computational methodologies, such as machine learning and predictive modeling, allows the team to delve deeper into the intricate labyrinth of microbial interactions. Fern’s prowess in AI is enabling the extraction of nuanced patterns and insights from voluminous datasets, translating them into coherent knowledge about microbial ecosystems’ structures, functions, and influences. David’s strengths in computational biology and AI are key to this effort, and her team has already made headway in applying natural language processing in the human microbiome dataset. These analytical frameworks are crucial to interpreting the data generated from metagenomic sequencing techniques and the multi-omics approaches employed in the study.
Impacts and implications
In addition to advancing scientific understanding, the microbiome project is developing innovative, open-source analytic tools that empower other scientists to contribute more effectively to the field. The project is also committed to fostering effective training, outreach, and assessment programs to nurture the future careers of microbiome scientists from diverse backgrounds. Ultimately, the work is anticipated to develop principles for predicting microbiome-host interactions in response to environmental change, and to yield foundational insights into how human activity impacts the environment through microbiomes. The expected result is significant advancement in the way we manage microbiomes, with profound implications for natural resource management, agriculture, and human health.
If you’re interested in connecting with the AI and Robotics Program for hiring and collaborative projects, please contact AI-OSU@oregonstate.edu.




